The Kendrick mass defect (KMD) is calculated by subtracting the Kendrick mass
(calc_km) from the nominal Kendrick mass
(calc_nominal_km). The the References section for
more information.
References
Edward Kendrick, Anal. Chem. 1963, 35, 2146–2154.
C. A. Hughey, C. L. Hendrickson, R. P. Rodgers, A. G. Marshall, K. Qian, Anal. Chem. 2001, 73, 4676–4681.
Examples
# Calculate the Kendrick mass defects for two measured masses with
# CH2 as the repeating unit.
# See Hughey et al. in the References section above
calc_kmd(c(351.3269, 365.3425))
#> [1] 0.06539637 0.06544638
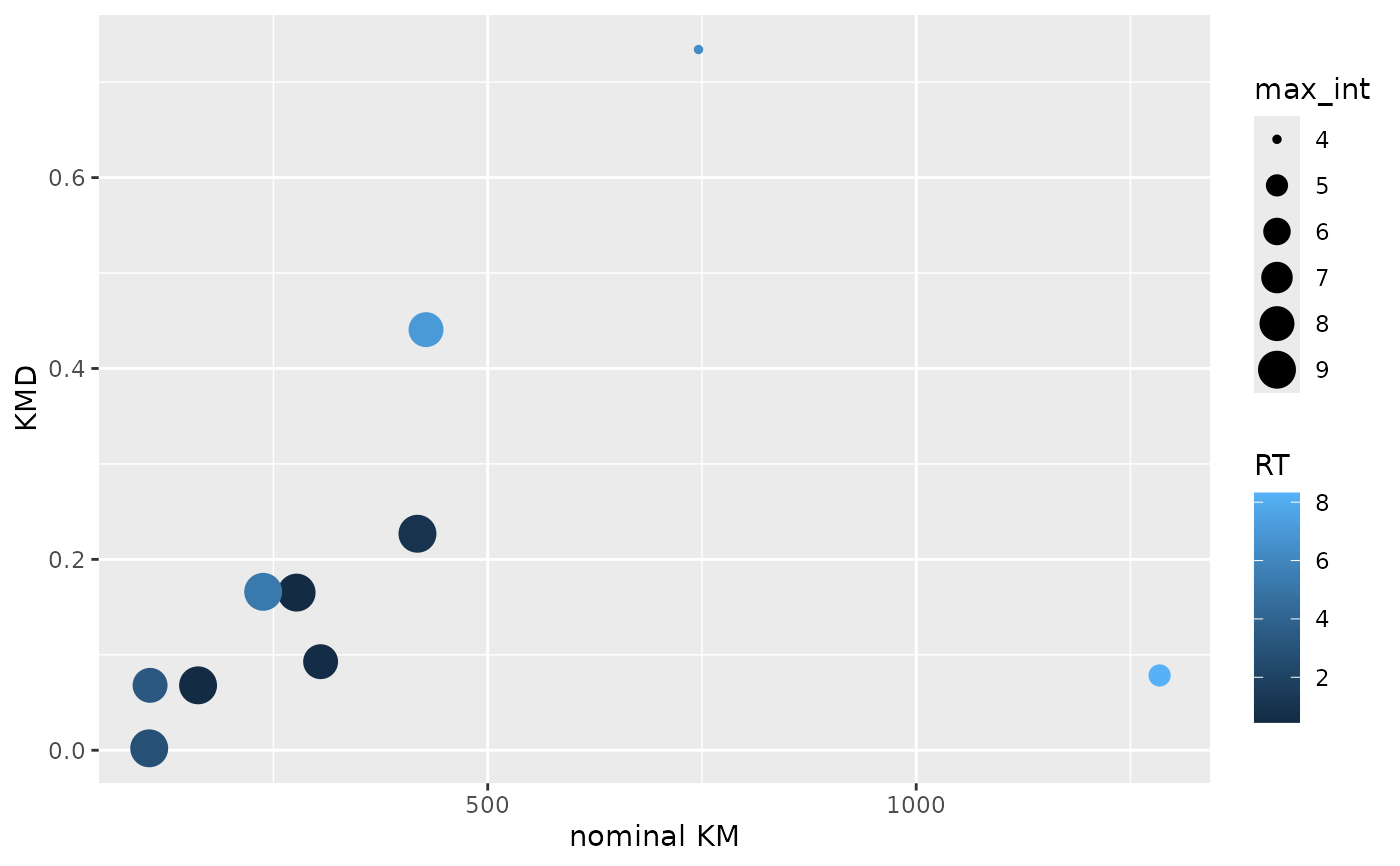
# Construct a KMD plot from m/z values.
# RT is mapped to color and the feature-wise maximum intensity to size.
toy_metaboscape %>%
dplyr::group_by(UID, `m/z`, RT) %>%
dplyr::summarise(max_int = max(Intensity, na.rm = TRUE)) %>%
dplyr::ungroup() %>%
dplyr::mutate(KMD = calc_kmd(`m/z`),
`nominal KM` = calc_nominal_km(`m/z`)) %>%
ggplot2::ggplot(ggplot2::aes(x = `nominal KM`,
y = KMD,
size = max_int,
color = RT)) +
ggplot2::geom_point()
#> `summarise()` has regrouped the output.
#> ℹ Summaries were computed grouped by UID, m/z, and RT.
#> ℹ Output is grouped by UID and m/z.
#> ℹ Use `summarise(.groups = "drop_last")` to silence this message.
#> ℹ Use `summarise(.by = c(UID, m/z, RT))` for per-operation grouping
#> (`?dplyr::dplyr_by`) instead.